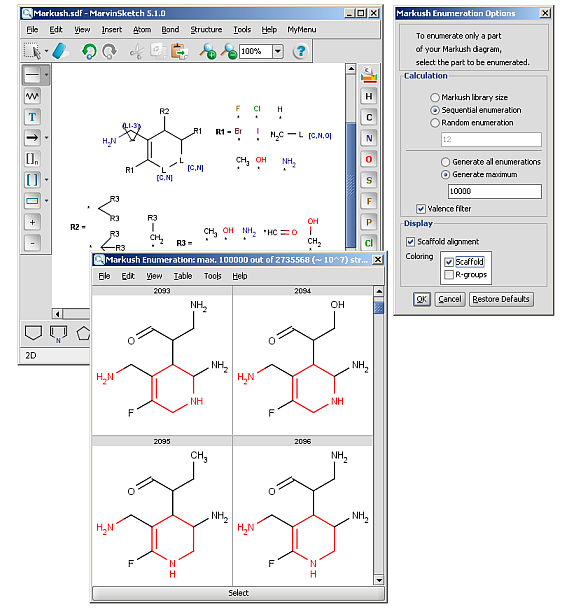
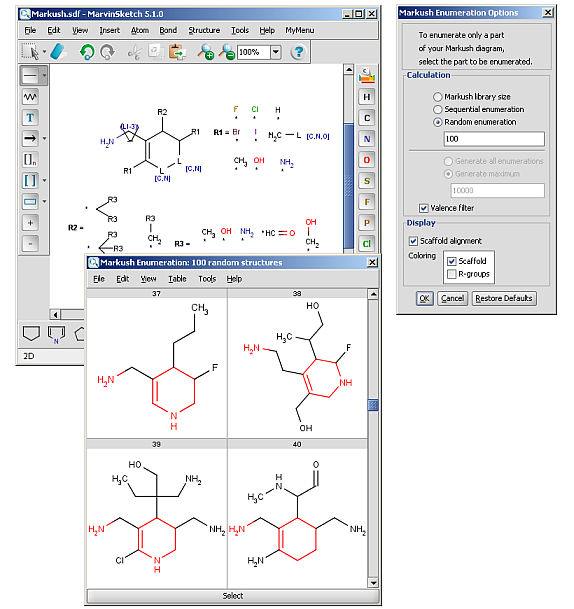
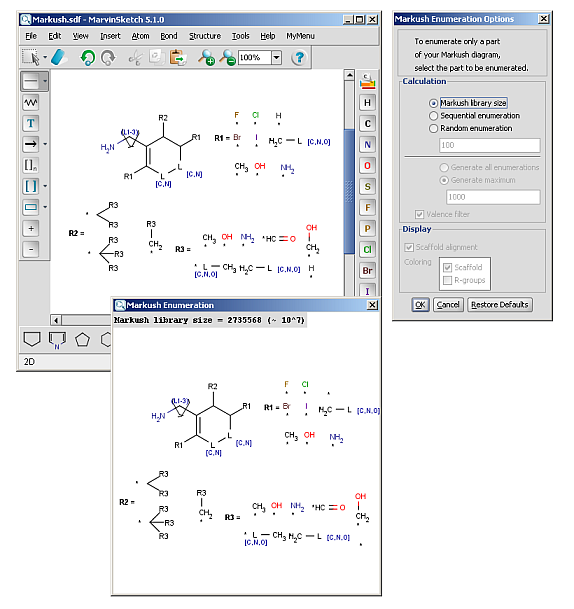
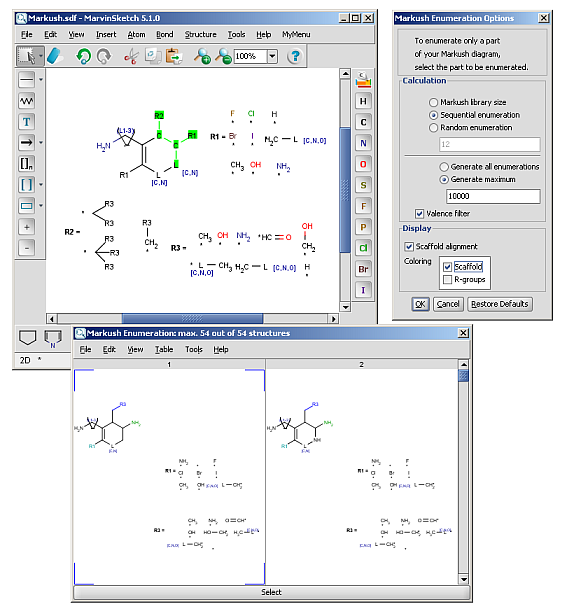
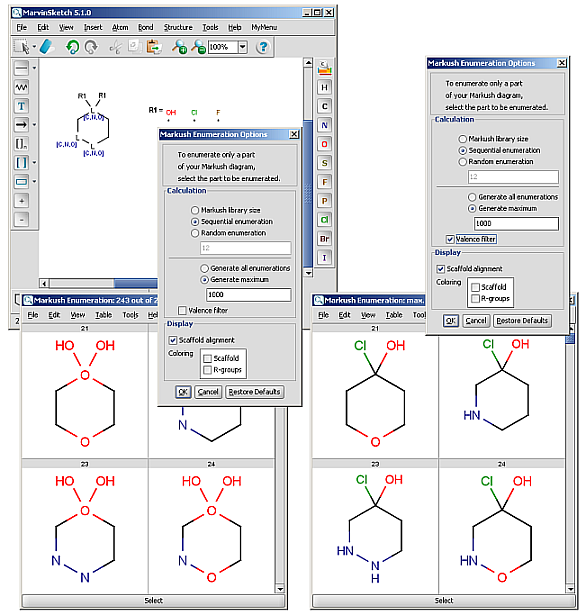
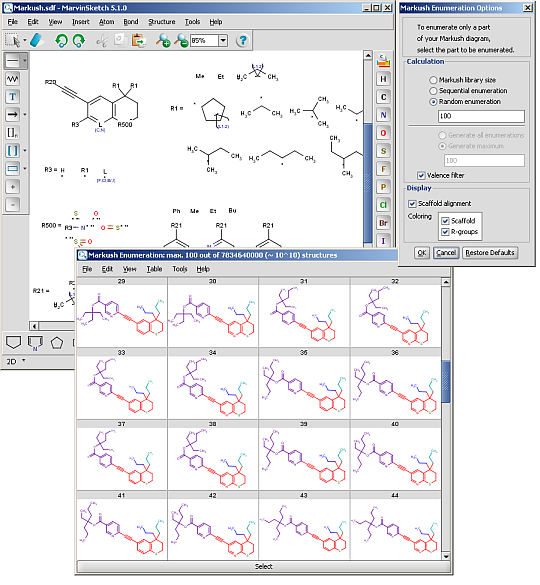
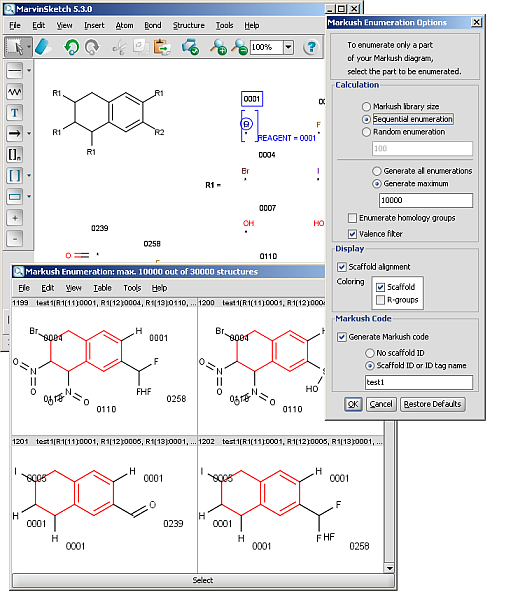
R-groups (also referred to as "substituent variation") are the most widely known Markush generic features. The variable part of the structure is denoted by an R-atom (eg. R1), and the definitions are given separately. In each definition the connection points must be defined to show where the bonds of the R-atom are linked. R-atoms can appear in both rings and chains and can have up to two attachments points. The same R-atom can appear multiple times, and the different occurrences are handled as different cases. (So they can be substituted with different definitions.) R-group nesting in R-group definitions is allowed to any depth, but without recursion. (An R-group definition cannot use the R-atom it is defining, not even through the use of other embedding R-atom(s).) R-groups up to number R32767 can be used.
| Example | Example Markush library member |
|---|---|
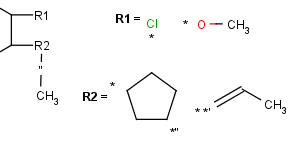 |
 |
R-group drawing in Marvin Sketch is described in the Marvin Sketch User's Guide.
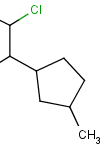
Atom lists are another example of substituent variation. They define lists of atom types at a given position. There is no restriction for the length of the list and for bond count of atom lists. Atom list drawing in Marvin Sketch is described here.
| Example | Example Markush library member |
|---|---|
 |
 |
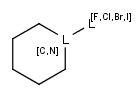
The following bond lists (generic bond types) are supported by the plugin: single or double, any(single, double or triple), single or aromatic, double or aromatic. In Marvin Sketch, bond lists are accessible amongst query bond types in the bonds pop-up menu.
| Example | Example Markush library member |
|---|---|
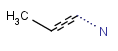 |
Link nodes are atoms that may repeat between two of their designated bonds (called outer bonds, denoted by brackets). All other substituents (if exist) repeat together with the atom. In the results, the new bonds between the repeating atoms will have the bond type of the lower order outer bond. Link nodes can be drawn in Marvin Sketch using the popup menu.
| Example | Example Markush library member |
|---|---|
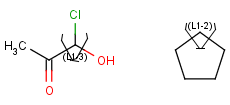 |
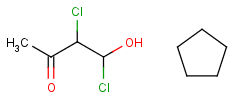 |
Repeating units represent structural parts that can be repeated several times. The repeating unit is enclosed in brackets with one or two head and the same number of tail crossing bonds. (Head crossing bonds go through the left bracket.) Two bond pairs represent ladder type repeating units. The repetition range is a comma-separated list of possible repetitions or repetition intervals, e.g. "1,3,5-9". The repetition pattern specifies the way how the subsequent repeated units are linked together: it can be head-to-head(hh), head-to-tail(ht) or either/unknown(eu) (the either/unknown case is not handled by the search software). In case of ladder type polymers there is also a flip(f) option that defines that the top and bottom crossing bonds are flipped during each connection. repeating groups with specified repetition ranges.
Repeating unit drawing is described in the Marvin Sketch Help here, and ladder-type bracket drawing is described at the polymer drawing section.
| Example | Example Markush library member |
|---|---|
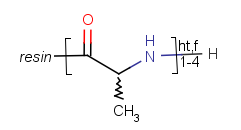
|
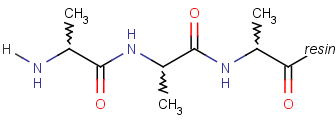
|
Position variation bonds are bonds attached to variable atoms at one or both end positions. The set of variable atoms is drawn as a multicenter group. A position variation bond connects one atom from one end position to one atom from the other end position. If the end position is a single atom then the bond is attached to this atom, if the end position is a multicenter group then the bond is attached to an arbitrary member of the group. Position variation drawing in Marvin Sketch is described in Help.
Limitations:
- Substructure search is not yet prepared to handle the case when both end positions are multicenter groups.
- A multicenter end position is not allowed to contain R-atoms.
- A multicenter end position is not allowed to contain another position variation bond (ie, position variation bonds cannot be nested).
If a link node is a member of a multicenter group then the group will include the repeated atoms as well in case when the original multicenter group contains no more atoms from the link fragment, otherwise the position variation bond is part of the link fragment and repeated together with the link node. Although an R-atom is not allowed to take part in position variation, it can be the single-atom end position of a position variation bond, in which case its attachment point is connected to the bond.
| Example | Example Markush library members |
|---|---|
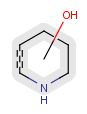
|
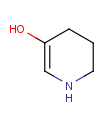
|
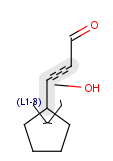
|
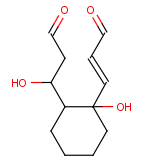
|
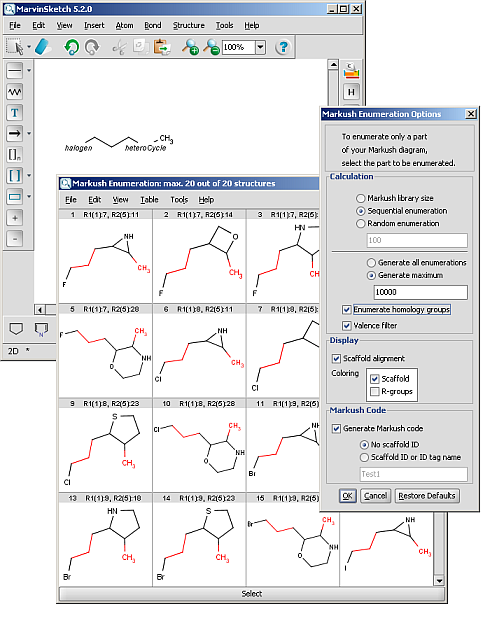
Homology groups stand for sets of homologous molecular parts (e.g. functional groups). These are represented by pseudo atoms labelled with the common chemical annotation of the groups (alkyl, aryl, heterocycle etc.). See the detailed definition of these groups in a separate document. The pseudo atoms can be most easily drawn in Marvin Sketch using the Homology Groups template group.
| Example | Example Markush library member |
|---|---|
|
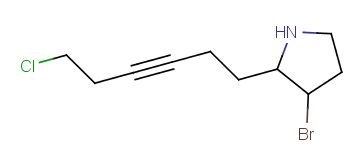
|
There are two major types of homology groups regarding their way of definition:
- Built-in groups are defined by specific structural properties of the group. These groups are not enumerated during searching, but the query structure is recognized as fulfilling the requirements for such a structure. The possible number of covered structures is usually infinite, unless the number of atoms is limited. Examples of built-in groups are alkyl, aryl, heterocycle, etc.
- User-defined groups are explicitly defined and only the listed structures can match on these homology groups. The definition is given in the form of an R-group definition, and any of the generic features discussed in this chapter can be used in the definition. These definitions can be customized by the user, and may be context-specific. (E.g. protecting group definition depends on which functional group it is protecting.)
Read more about homology groups.