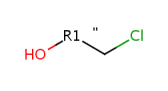
R-atom attachment order is determined by the neighbor atom index order, the second order neighbor
is marked with a " on the attachment bond:
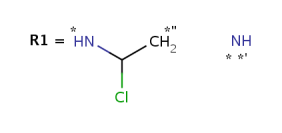
Attachment points in R-group definitions are marked with * and *" signs:
Codename: vmn
Markush structure files complying the Markush DARC format are processed by Marvin, though with some
limitations. Interpretation of the main VMN features are
listed below. Multiple (more than 2) attachment R-groups
are temporarily represented by attached data, as described below.
This representation will be changed to its final version for the next major release.
Peptide connection bonds may not be processed correctly and
some peptides are not supported yet.
The AMN file is looked for in the directory of
the VMN file, with the same name and .amn extension
(e.g. the AMN file of 46mrk001.vmn is 46mrk001.amn).
The AMN attributes and some of the atom attributes
are not processed by search and enumeration but displayed in atom labels.
G0 is read in as root structure while G1, G2, ...
are stored in corresponding R-groups R1, R2, ... The representation of
attachments is described below.10 elements in a repetition range.100) on the atoms of the repeating unit, with the appropriate repetition range text recorded in the corresponding AMN file (e.g. M100=1-4). Note, that there is a limitation on the number of elements in a repetition range (at most 10) and on the number of crossing bonds that can be processed (2 or 4).The following structure shortcuts (abbreviated groups) are supported:
| ACE | BU | C2, C3, ..., C20 | CN | CO1 | CO2 |
| COI | ET | IBU | IPR | MBE | NBU |
| NO2 | NPR | OBE | PBE | PH | PO3 |
| PO4 | SBU | SO2 | SO3 | TBU |
The following amino acids (peptide abbreviated groups) are supported:
| ALA | ARG | ASN | ASP | CYS | GLN |
| GLU | GLY | HIS | ILE | LEU | LYS |
| MET | PHE | PRO | SER | THR | TRY |
| TYR | VAL |
For more information, refer to the Peptide import documentation.
Superatoms representing homology groups are read in as pseudo atoms. The following homologies are interpreted by enumeration and search:
| CHK | CHE | CHY | CYC | ARY | HET |
| HEA | HEF | UNK | MX | AMX | A35 |
| TRM | LAN | ACT | HAL | ACY | PRT |
| XX |
For a detailed description of the interpretation, refer to the Homology groups in Markush structures manual.
Note, that the current representation is temporary and will be replaced by a final solution in the next major release.
The usual attachment point representation can be used for 1- and 2-attachment R-groups:
R-atom attachment order is determined by the neighbor atom index order, the second order neighbor
is marked with a " on the attachment bond:

Attachment points in R-group definitions are marked with * and *" signs:

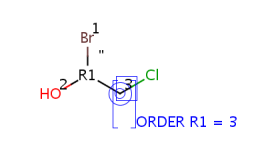
For R-groups with more than 2 attachments we use the following temporary representation:
R-atom attachment order numbers is stored in ORDER R<n> attached data fields,
where n is the R-group ID (e.g. ORDER R1 for R1,
ORDER R2 for R2, etc.):
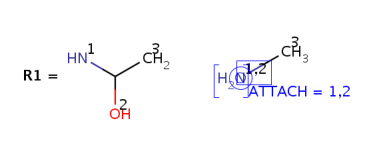
Attachment point numbers in R-group definitions are stored in the ATTACH attached
data field:
Attached data can also be set manually in MarvinSketch, as described in the MarvinSketch Chemical Features manual.
Both the attachment order numbers and the attachment point numbers are consecutive positive integers
1, 2, 3, ..., where attachment i is to be connected to the R-atom neighbor
with order number i, for each i. Multiple attachment numbers on the same atom
are separated by commas.
Attachment point numbers can be omitted for single atom R-group definition members. If the order numbers are missing for an R-atom, then the atom index order of the neighboring atoms is used to determine the order numbers.
The number of elements in a repetition range is limited to 10 (e.g. range M100=2,5- is interpreted as M100=2,5-13). Repeating units with more than 4 crossing bonds are not processed by search and enumeration.
The following peptides are not supported by Peptide import (and therefore are not read in from VMN files):
| ABU | aminobutyric acid |
| ASU | aminosuberic acid |
| GLP | pyroglumatic acid |
| HCY | homocysteine |
| HSE | homoserine |
| NLE | norleucine |
| NVA | norvaline |
| ORN | ornithine |
| SAR | sarcosine |
| STA | statine |
Note, that peptide connection bonds are not handled currently, therefore peptide sequences may not be correct.
The following superatoms (homologies) are not supported by search and enumeration (but read in and displayed as pseudo-atoms):
| POL | PEG | XX | DYE | PRT |
The detailed description of homology interpretation is described in the Homology groups in Markush structures manual.
The following atom attributes are not processed by search and enumeration (but displayed in atom labels):
| CR | Carbon chain/ring attribute |
| DT | Deuterium and tritium count |
| MU | Multiplier attribute |
| NU | Numerotation attribute (link to AMN data) |
| PA | Polymer indicator |
| SP | Position indicator |
VMN export is not available yet.